Protein Binding and Interaction Analysis of Human Pathogen Protein Targets
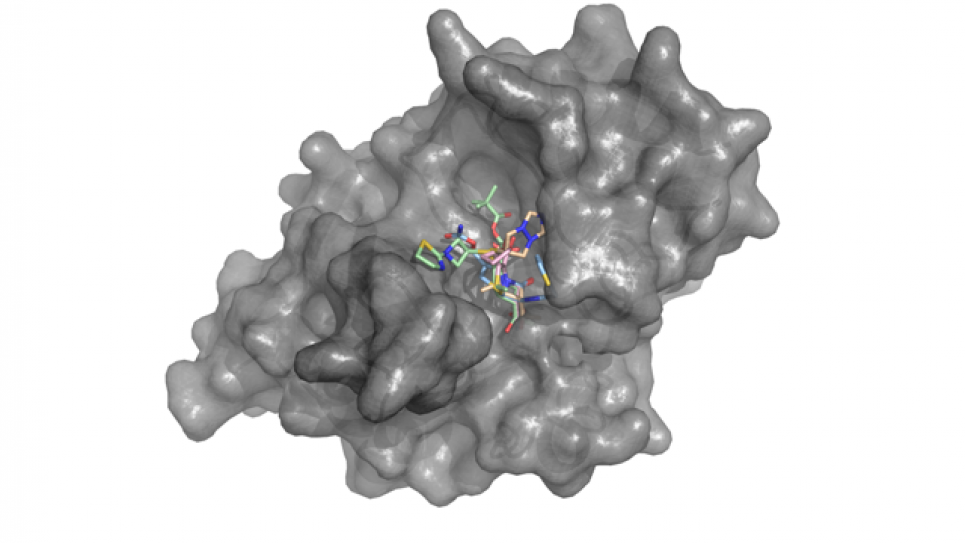
This project is performing computational discovery studies of high-value biomedical targets of human pathogens, bioterrorism agents and human disease related proteins identified by the Midwest Center for Structural Genomics (MCSG; Argonne National Laboratory) and the Center for Structural Genomics of Infectious Diseases (CGSID; Northwestern University). The research team is applying the novel Free Energy Perturbation Replica-Exchange Molecular Dynamics with respect to the thermodynamic parameter λ (FEP/λ-REMD) method to
conduct unprecedented large-scale studies to evaluate and benchmark the higher order proteinligand binding free energy calculations. They are also applying parallel/parallel accelerated FEP algorithm (implemented on greatly scalable software NAMD) to realize high throughput compounds screening on Blue Gene P and Q. The computational results will be experimentally verified on select protein targets using inhibition assays and crystallographic techniques in collaboration with experimentalists at the MCSG and CSGID.
In addition to continued work on IMPDH, they will focus considerable focused on identifying inhibitors against New Delhi Metallo-β-lactamase (NDM-1). The imminent threat posed by the recent discovery and dissemination of the plasmid NDM-1 gene harbored by multiple pathogenic
microorganisms presents a global healthcare threat.