Studies of Large Conformational Changes in Biomolecular Machines
In biology, proteins, nucleic acids, and carbohydrates can be considered complex “molecular machines” that consume energy in order to perform specific biological functions. Their concerted actions trigger many of the critical activities that occur in living cells. In particular, membrane-associated proteins play essential roles in controlling the bidirectional flow of material and information. These proteins change shape and go through many conformational states to perform their function. Malfunction of some vital proteins can lead to diseases such as cancer.
To understand how membrane proteins operate and how they are impacted by disease, researchers need detailed knowledge about all the relevant conformational states, as well as the free energy changes that connect them. This INCITE project is aimed at gaining a deep mechanistic perspective of membrane protein function, linking structure to dynamics, by characterizing the free energy landscape that governs functional motions.
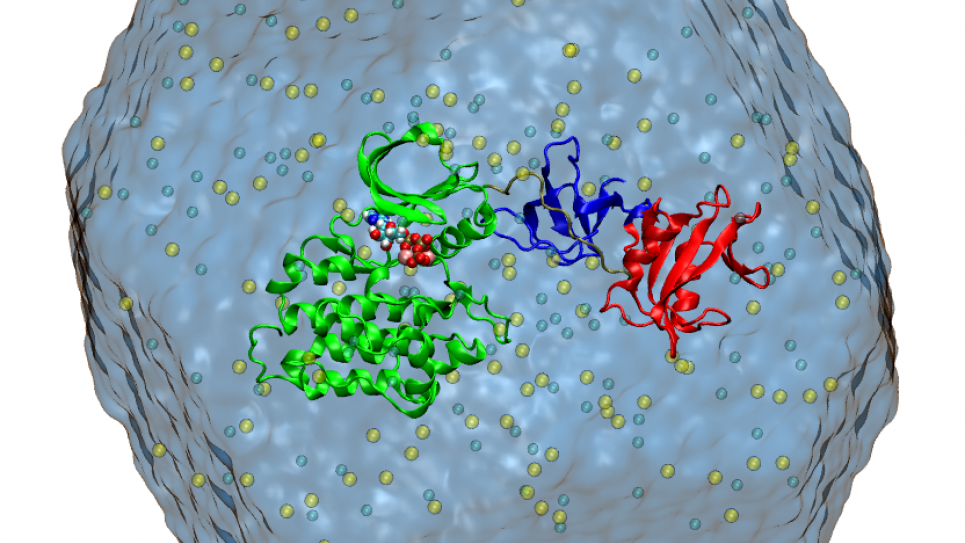
Within the unified computational perspective provided by free energy landscapes, two membrane proteins of increasing complexity and size are considered (a Src kinase and a P-type ionic pump). For this project, researchers will leverage NAMD/Charm++ with MPI-level multiple copy algorithms, a computational methodology that achieves extreme scalability on leadership-class supercomputers and is at the forefront in the field of biomolecular simulations.
By studying experimentally well-characterized systems of increasing size and complexity within a unified theoretical framework based on free energy landscapes, the researchers will push the envelope and advance the theory-modeling-simulation (TMS) technology. TMS offers a virtual route to address fundamental biological questions and to help solve the problem of rational protein design. The computations planned for this study will serve as a roadmap for simulating, visualizing, and elucidating how biomolecular nano-machines work.