Large-Scale Condensed Matter and Fluid Dynamics Simulations in Three Diverse Areas: Part II: Molecular Dynamics Study of Clay-Polymer Nanocomposites
We use large-scale molecular dynamics simulations combined with rare event sampling techniques to understand the unique chemical and physical aspects associated with clay-polymer-based nanocomposite materials. Tailoring the clay structure in polymers on the nanometer scale produces composites with novel material properties that have already been applied in numerous commercial applications. Once fundamentally understood, clay-polymer nanocomposites have the potential to become as widespread as traditional, conventional composite materials.
Scientific Approach
Researchers will perform non-equilibrium molecular dynamics (NEMD) simulations of clay-polymer nanocomposites, from which they will calculate material properties. The scale of the systems involved allows, for the first time, study of the arrangement of discrete platelets in atomistic detail.
Using a parallel tempering technique (replica exchange), scientists will simulate polymer penetration into the clay platelet, a process that occurs on a time scale outside that possible with a single molecular dynamics simulation. A greater understanding of the mechanism of intercalation in these materials will lead to improved techniques for nanocomposite formation.
Thirdly, researchers will generate coarse-grained representations of the clay platelets using the atomistic simulations of interaction parameters between the coarse-grained superatoms. Coarse-grained simulations are expected to be achieved in micro- to milliseconds.
Results
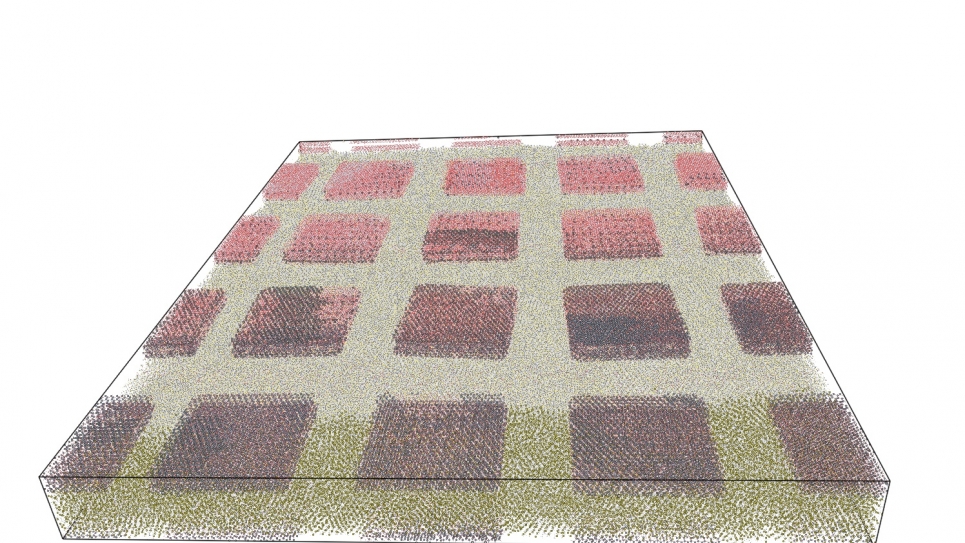
To date, scientists have created clay-polymer nanocomposite systems consisting of approximately 10 million, containing 16 isolated clay platelets–the first time such simulations of isolated platelets have been run. They have performed molecular dynamics simulations using the LAMMPS code up to a simulation time of 1ns, which is currently being extended, and will form the basis of elastic calculations to further understand the properties of clay-polymer nanocomposites. A smaller system consisting of a single platelet immersed in water allows researchers to understand the process of intercalation of small molecules into the interlayer spacing between clay sheets. It will be the basis for the parallel tempering technique, which will be used to study similar, but much rarer, intercalation of large polymer molecules.
Scientists have also developed a protocol for building coarse-grained simulation from all-atom simulations, from which we have built a test system of 500,000 coarse-grained particles, which corresponds to a 4-million-atom system.
Future Efforts
Moving forward, researchers will perform NEMD simulations of the clay-polymer nanocomposites, from which they will calculate material properties, including the bending modulus, Young’s modulus, and Poisson’s ratio. The objective is a greater understanding of the enhancement mechanism, which may be a cooperative effect over the many disparate length scales. They will also be able to study the interactions between separate platelets, which may determine the rheology of clay nanocomposites. In addition, scientists will ascertain how the long-range order and arrangement of the platelets affect the material properties of the nanocomposites. Because of the sheer scale involved, no previous study has ever considered the arrangement of discrete platelets in atomistic detail.
Their coarse-grained simulations will allow exploration of greater time-scales than possible with atomistic molecular dynamics, providing another avenue to explore the intercalation mechanism and long-range order. From the biological domain, researchers expect their coarse-grained simulations to allow speed-ups of many orders of magnitude. Further, they expect their coarse-grained simulations to be achieved in micro- to milliseconds, approaching realistic synthesis time scales.