Molecular Engineering through Free Energy Mapping
Block polymers (BP) are known to self-assemble into a wide variety of nanostructured morphologies, as do liquid crystals (LC) which similarly adopt ordered morphologies capable of interacting with light in distinct ways. These are examples of mesoscale assembly, where the behavior of a material over relatively long length-scales emerges from the subtle balance of disparate molecular-level interactions.
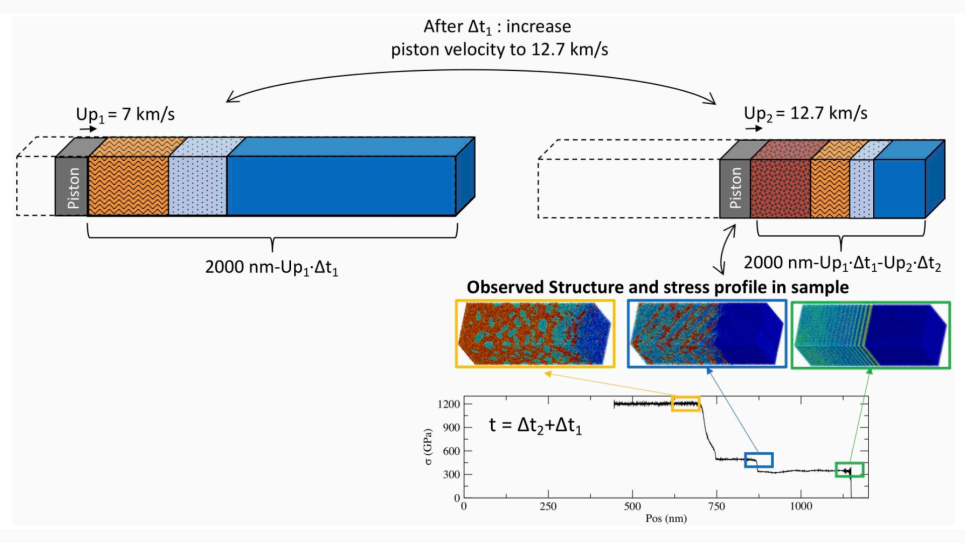
Yet, key questions and phenomena arising at the mesoscale remain unanswered, and can only be addressed by simulation over time and length scales that, until now, were inaccessible. This project seeks to demonstrate the predictive ability of powerful new models and methods that will utilize the large-scale computing power of ALCF’s Mira, an IBM Blue Gene/Q. A team of researchers from the Institute for Molecular Engineering, University of Chicago and Argonne National Laboratory, will use these tools to identify mesoscale assemblies in atomistic and mesoscale simulations, and map their free energies with unprecedented accuracy through simulations of large ensembles.
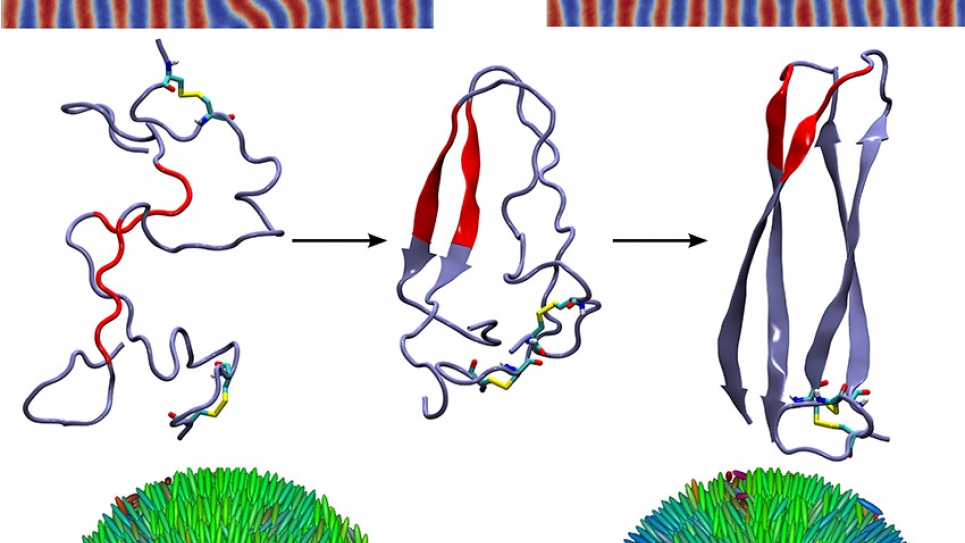
Focusing on three platforms, this INCITE research will examine the dynamics of directed BP assembly, which is of considerable importance in the fabrication of next-generation integrated circuits and high-density storage media. The formation of polypeptide aggregates—which are implicated in several neurodegenerative diseases and known to be extremely toxic—will be examined, as will the formation of defects in LC systems, and the use of those systems in biosensors and programmed nanoparticle assembly.
Each of these platforms builds on the research group’s expertise in novel free-energy calculations and is of considerable technological importance. The results could provide wideranging application in the design of microelectronics, self-assembly, and human health.