Simulation and modeling of membranes interactions with unstructured proteins and computational design of membrane channels for absorption of specified ions
Researchers led by Igor Tsigelny from the University of California–San Diego (UCSD) are seeking to understand why some proteins aggregate and what structural or sequence features may increase their propensity to do so, how proteins penetrate membranes to form pore-like structures, and what role protective molecules play in influencing protein aggregation. The findings of this study offer important implications for identifying the molecular basis of Parkinson’s disease and effective ways to treat it.
Through modeling on Intrepid, the IBM Blue Gene/P supercomputer at the Argonne Leadership Computing Facility, the team has produced three drug candidates with high potential for treating the disease by stopping or preventing pore formation. Each theoretical design was tested in the laboratory and found to be effective. Patents for all three drug candidates are pending.
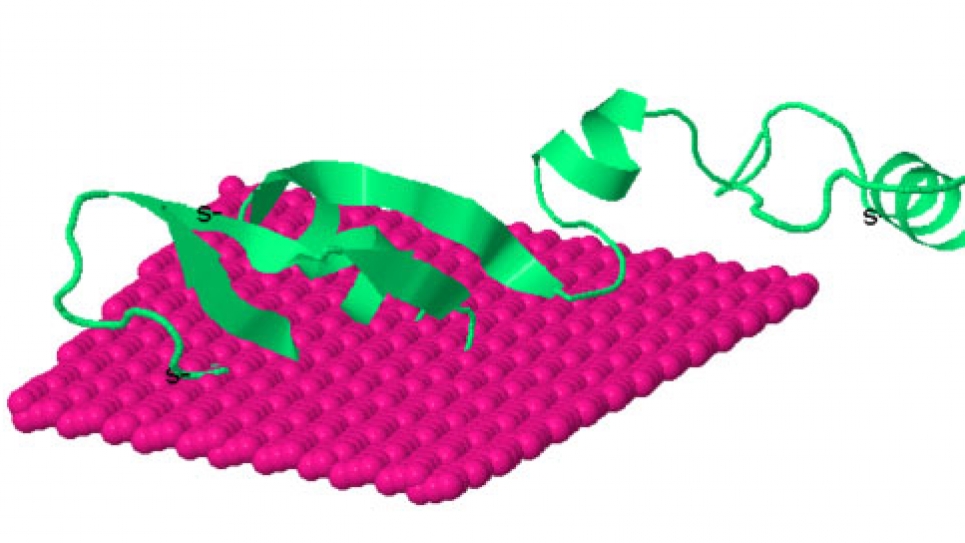
Simulations conducted by the UCSD scientists also have shown the clear creation of annular protein aggregates that can penetrate to the bacterial membranes and create the pores in them. Such pores are open for absorption of radionuclides. The team’s exploration of pore formation in bacterial membranes may also yield answers to problems associated with environmental waste by reducing radionuclides—atoms that emit harmful radioactivity—and eliminating environmental waste through absorption. Working with scientists in Ukraine, the team is organizing an initiative in which it will conduct experimental validation of efforts to clean radionuclides from water in Chernobyl.