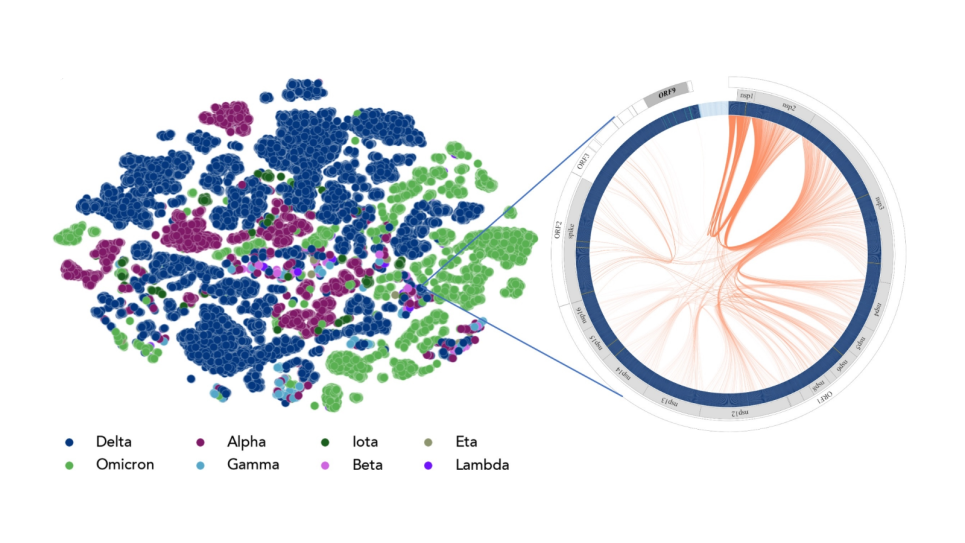
Genome-scale language models (GenSLMs) for predictive molecular epidemiology.
This project aims to build foundation models for large-scale genomic datasets for continuous monitoring and tracking of pathogens. It will thus increase biopreparedness and will benefit the community by making GenSLM models, data, and code available to a broad user base, who can fine-tune the foundation models for their own downstream predictive tasks.
The potential for extant and emerging pathogens to become global health crises necessitates the development of novel methods for proactively engaging these threats before they become pandemic. Recent advances in machine learning and artificial intelligence—specifically, large language models (LLMs)—provide powerful tools for predictive modeling and monitoring of pathogens of concern. The team’s prior work developing Genome-scale Language Models (GenSLMs) demonstrated the potential for LLMs to predict future SARS-CoV-2 variants of concern prior to their emergence by modeling the evolutionary process. In this project the team builds on that work by scaling GenSLMs beyond the (relatively) simple SARS-CoV-2 to multi-segmented viruses and comparatively enormous bacterial genomes, and even further to more complex eukaryotic organisms including yeast and humans.
This project will thus increase biopreparedness by providing a continuous watchlist of pandemic-potential variants across several different pathogens; and will additionally benefit the community by making GenSLM models, data, and code available to a broad user base, who can fine-tune our foundation models for their own downstream predictive tasks.