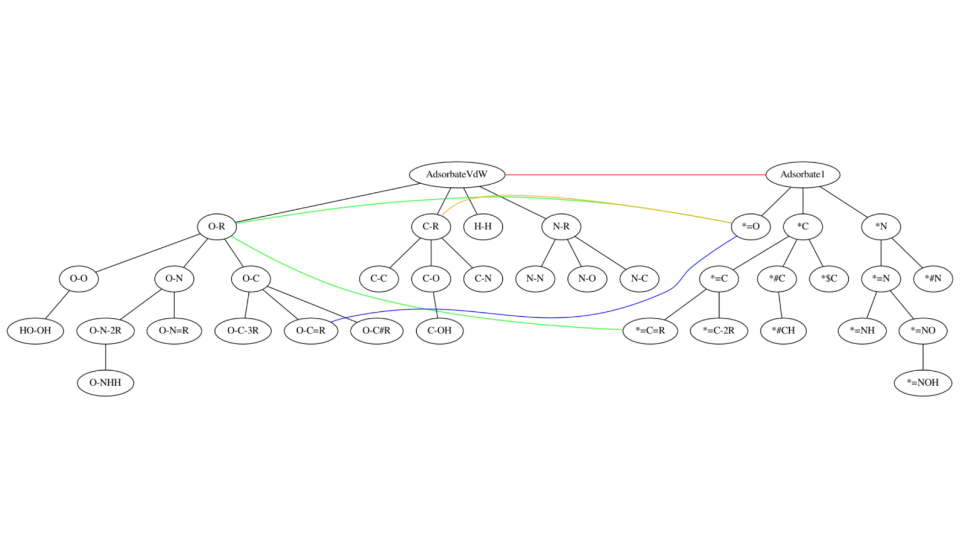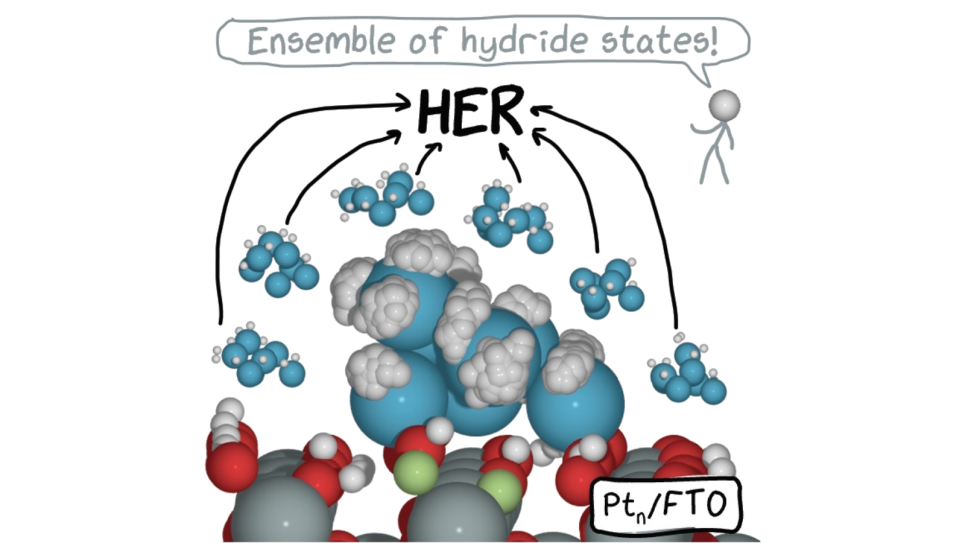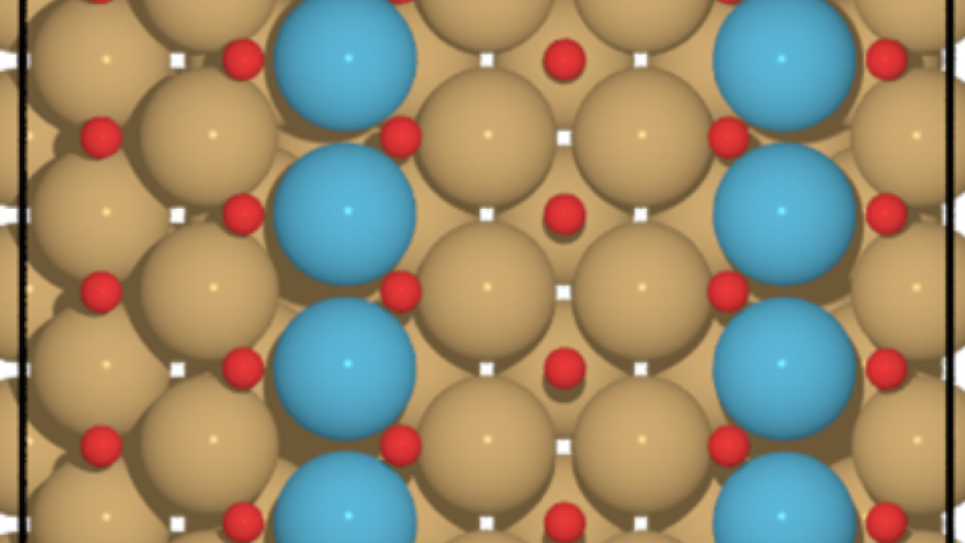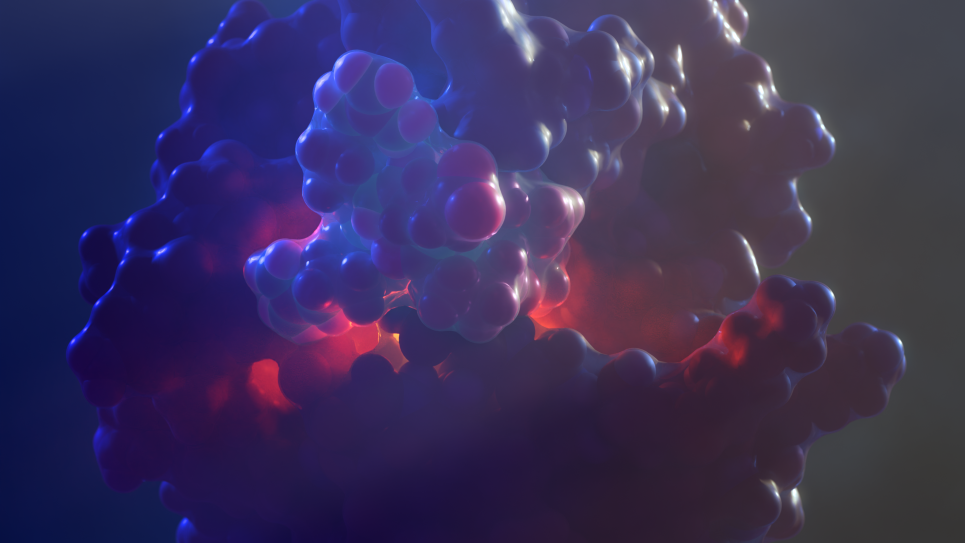
X-ray crystal structure of a computationally-designed peptide macrocycle inhibitor, NDM1i-1G (nicknamed "Moriarty"), bound to the New Delhi metallo-β-lactamase 1 (NDM-1), an antibiotic resistance factor. The inhibitor, highlighted in pale green-blue, binds to and occludes the active site of the enzyme, shown in orange, preventing the enzyme from breaking down penicillin and other related antibiotics. Simulations performed on DOE supercomputers allowed the ranking of candidate designs, permitting the discovery of this molecule from a screen of only seven molecules that were synthesized in the chemical laboratory. In contrast, conventional drug discovery efforts that are not bolstered by computational methods require the screening of hundreds of thousands to millions of compounds to find an initial hit. Image: Vikram K. Mulligan, Flatiron Institute.