Computational Studies of Nucleosome Stability
Scientists at Argonne are studying the mechanical properties of DNA. Modeling these properties is now feasible by conducting simulations on the ALCF’s supercomputer. By understanding the nucleosomes, which account for 75-90% of the packaging of genomic DNA in mammalian cells, researchers can use this information to identify abnormalities that can be used as biomarkers in the diagnosis of genetic diseases.
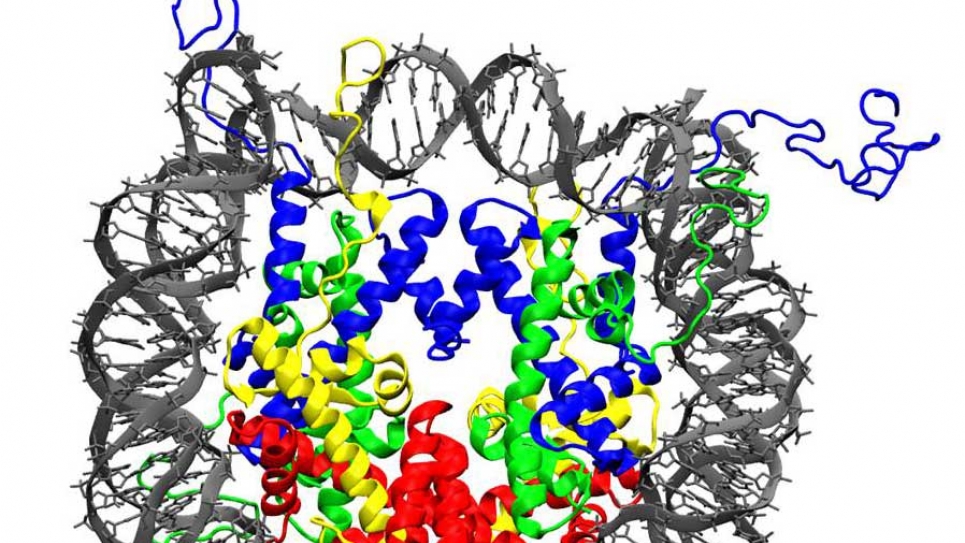
This INCITE research project led by George Schatz is concerned with the molecular dynamics and electronic structure studies of the nucleosomes, which are complexes of DNA and proteins in chromatin and account for 75-90% of the packaging of the genomic DNA in mammalian cells. Nucleosomes consist of 147 base pair (bp) of DNA wrapped 1.7 times around a complex of proteins known as the histone octamer to give an overall nucleosome diameter of 11 nm, and a rather small DNA radius of curvature of 4.1 nm. Understanding the base-pair sequence dependence and propensities formation has long been a goal in cell biology as nucleosome stability is thought to be an important component of transcriptional regulation.
The research team will make a fundamental advance in computational modeling of nucleosome stability using the Argonne Leadership Computing Facility. It is clear that there is now a significant opportunity for taking a major step forward in our understanding of nucleosome binding through the use of high performance computation (HPC). Empirical potential classical molecular dynamics calculations are now possible for whole nucleosome structures, and such calculations on an HPC system for a long enough time (~1 microsecond) to equilibrate structures that include explicit water and ions would enable us to determine useful information about nucleosome binding, including both sequence dependence effects and the effect of epigenetic markers. Electronic-structure studies of smaller models of the nucleosomes using the fragment molecular orbital (FMO) method will also be performed.