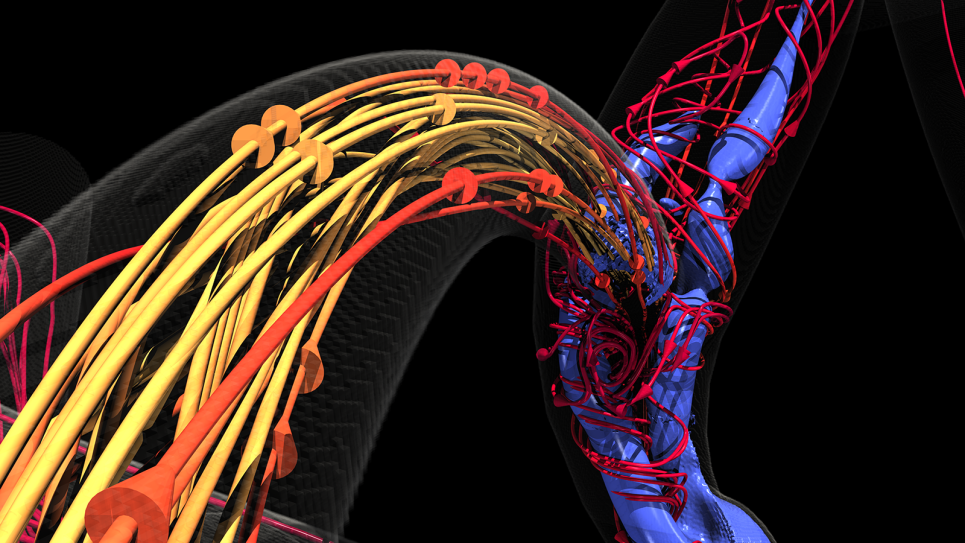
Several years ago, Dr. Amanda Randles, Alfred Winborne Mordecai, and Victoria Stover Mordecai Associate Professor Department of Biomedical Engineering at Duke University, and her team developed a computational fluid dynamics model called HARVEY that predicts and simulates how blood cells flow through the human body. To put the scale of that endeavor in context, consider this: an average person’s vascular system has enough plumbing to stretch from Seattle to Tokyo and back again. Some capillaries are so tiny that blood cells must move single file.
The researchers have come a long way since then. The scientists now focus on a different goal — adapting the HARVEY model for cancer research. Using HARVEY simulations, the team hopes to identify likely locations where a metastasized cancer cell might lodge in the body after breaking away from a primary tumor. When successful, Dr. Randles hopes the simulations can help oncologists focus their scrutiny on areas where secondary tumors could form. If a tumor does not seed in those locations, metastasized cancer cells are much less likely to implant and grow elsewhere.
An enormous challenge
The complexity of HARVEY simulations and the petabytes of data involved with them require the capabilities of advanced technologies supporting the Aurora exascale-capable supercomputer at The Department of Energy’s (DoE) Argonne National Laboratory (ANL). Aurora will include over 10,000 blades supported by 20,000 52-core Intel Xeon Max Series CPUs with 128 MB of high bandwidth memory each, 60,000 Intel Data Center GPU Max Series, Distributed Asynchronous Object Storage (DAOS), and more.
In modeling and simulations of this complexity, the HARVEY team needed to break their work into parts. “Some of our team worked on in-situ visualization and machine learning training within HARVEY. Others spent a lot of time with Intel and the team at ANL to determine the optimal programming model,” said Dr. Randles. Toward the latter endeavor, Dr. Randles’ team, in collaboration with Dr. John Gounley at ORNL, created a proxy application and performance model that simulates HARVEY’s performance expectations and helps identify the ideal programming models.
When visualizing terabytes of data like HARVEY does, the rendering component alone can require a supercomputer. Dr. Randles noted, “There are around 35 trillion blood cells in the body. In 2017, we could only track a single cell, but today, we regularly simulate regions with 100 million cells. What we can do now shows how far HPC technology advanced in just a few years. Our work can involve so much data that we can’t visualize it without some kind of framework to run the visualization in parallel. It’s a whole different scope of problem to solve technically.” Aurora will provide more than enough horsepower to handle the entire workload when it comes online.
According to Dr. Randles, different types of cancer cells can vary in size or stiffness of their cell walls. Even small factors like these can compound the HARVEY challenge since models must accommodate atypical cells moving through the vascular system and how they interact with normal cells.
To overcome this challenge, the team works closely with experimental labs to ensure models accurately mimic that cell type’s behavior. Dr. Randles detailed the difficulty. “We work with a great team at Dr. Lydia Sohn’s lab in Berkeley that uses an in-vitro microfluidic device to measure the cancer cell properties. We may not be able to measure the cell’s elasticity, but if we make a digital twin with their model, we can match their experiment. That allows us to work backward and determine how to tweak our modeling parameters to match their experimental data. From there, we can couple the experimental data with the computational model to measure factors that are impossible to do directly.”
Optimizing for Aurora
While readying HARVEY code for its valuable time on Aurora, the team benefitted substantially from the open, oneAPI programming model. “I was amazed that a graduate student completed the huge task of preparing our code in just a couple weeks with the help of oneAPI. I’m not talking about our proxy model – I mean preparing HARVEY itself!” Added Dr. Randles, “The work required some manual interaction, but performance, even without tuning, was quite good.”
However, maximizing performance is a team effort. Dr. Randles said, “Intel helped us with profiling tools and identifying pain points like compiler flags. With resulting optimizations in place, we saw a big performance benefit we might have missed. Working with Intel helps us ensure we use Aurora’s underlying hardware to its full potential.”
While the team unleashed some of HARVEY’s potential on other supercomputers in the past, bottlenecks existed. Training HARVEY involves petabytes of data for every single blood cell, plus millions of time steps. Therefore, the model typically uses most system memory on even the largest HPC systems. Aurora will take their work a big step further. “While we can do in situ visualization now, the Intel Max GPUs and ray tracing capability will allow us to employ OSPRay to obtain photo-realistic visualization.”
Enormous potential
While the team’s first challenge was forming a generic vascular model, their long-term goal is far more profound. Eventually, HARVEY will have the ability to create a personalized model for individual patients. But that goal remains on the horizon. “It’s not necessarily a computing constraint anymore, said Dr. Randles. “We don’t have a full body scan of patients that reaches the level of resolution that allows us to understand a cancer cell’s interaction in the bloodstream. We’ll get there someday.”
While significant work remains, Dr. Randles and her team’s endeavor has enormous potential for medical applications. As Dr. Randles put it, “Right now, we see no major hurdles that would impede HARVEY’s eventual use in the fight against cancer. We’re confident we can scale on Aurora, and we’re very eager to see it help cancer patients live longer and healthier lives.”
Continued Dr. Randles, “I owe enormous thanks to researchers at ANL, including Joe Insley, Geng Liu, Victor Mateevitsi, Silvio Rizzi, Saumil Patel, and graduate students Aristotle Martin, William Ladd, and Ayman Yousef, who work in my lab. With all their help, we’ve advanced HARVEY’s original incarnation to a level we could only dream of five years ago.”
# # #
This article was produced as part of Intel’s editorial program, with the goal of highlighting cutting-edge science, research and innovation driven by the HPC and AI communities through advanced technology. The publisher of the content has final editing rights and determines what articles are published.