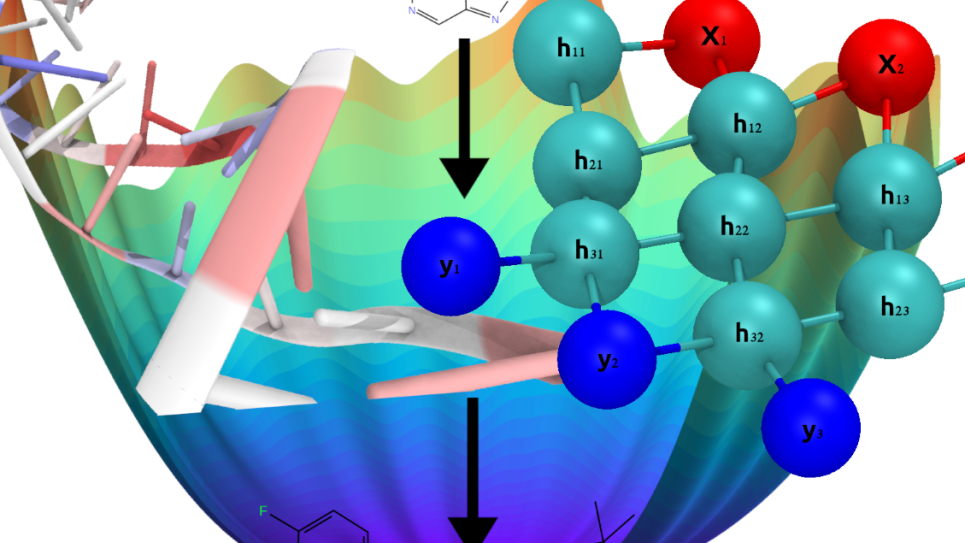
An artistic representation of the IMPECCABLE workflow that constructively combines physics-based simulations and machine learning approaches to accelerate the process of compound screening in drug discovery.
Coupling machine learning and physics-based methods, with this work researchers aim to accelerate the slow process of drug discovery, which typically lasts many years and costs billions of dollars—a major weakness in public health emergencies.
Wi this INCITE project, these researchers aim to accelerate the often slow process of drug discovery. Furthermore, virtual screening methods employed in drug discovery are currently hampered by their reliance on human intelligence in the application of chemical knowledge.
To overcome this, this team has developed a method called “IMPECCABLE” that involves sampling candidate compounds from both a billion-compound, synthetically accessible space, as well as from the output of a deep learning generative algorithm. It is an iterative method that loops its various modules consisting of a variety of physics-based scoring methods with increasing levels of accuracy as well as DL methods actively interacting with each other and becoming progressively more accurate in their predictions. The selected compounds are scored based on calculated binding free energies and fed back into the deep learning algorithm to iteratively refine predictive capability.
The approach this team takes will have direct applicability in the pharmaceutical industry for quick identification of potent binders for a given target protein and binding pocket. Using “IMPECCABLE,” the team has been able to analyze several million compounds from a set of orderable compound libraries to discover an inhibitor for the main protease of SARS-CoV2.