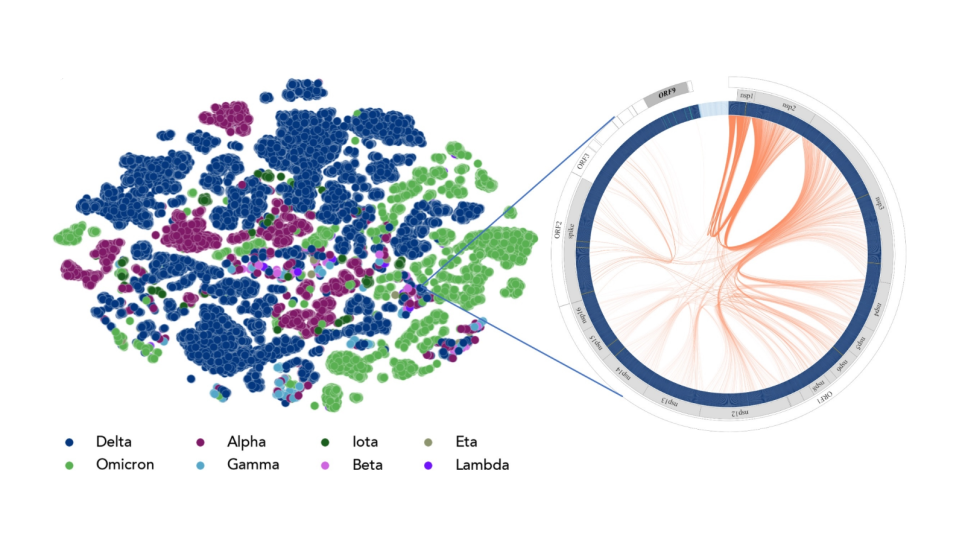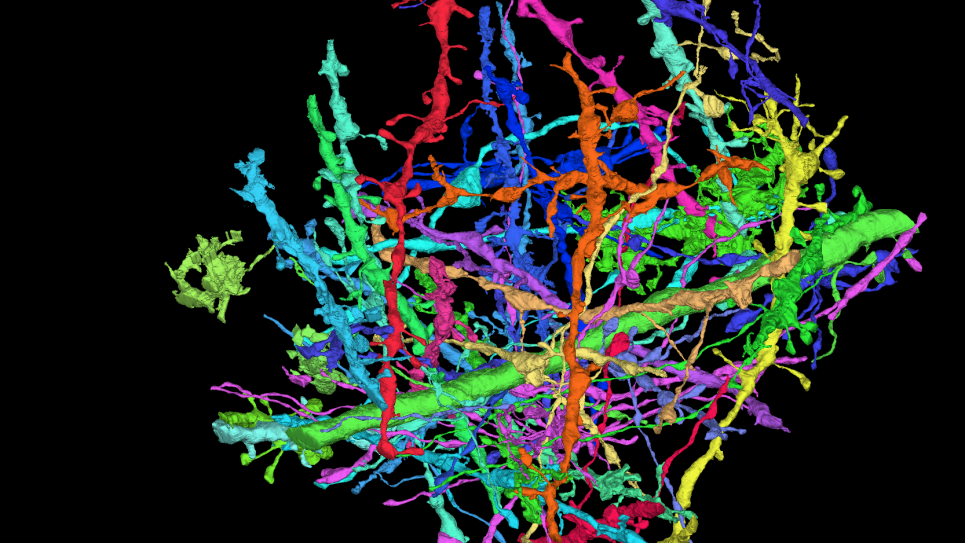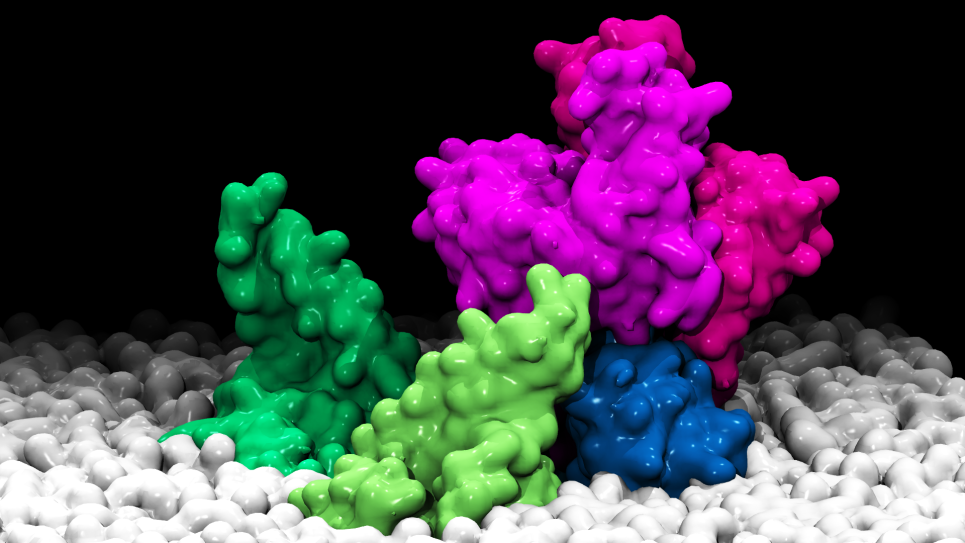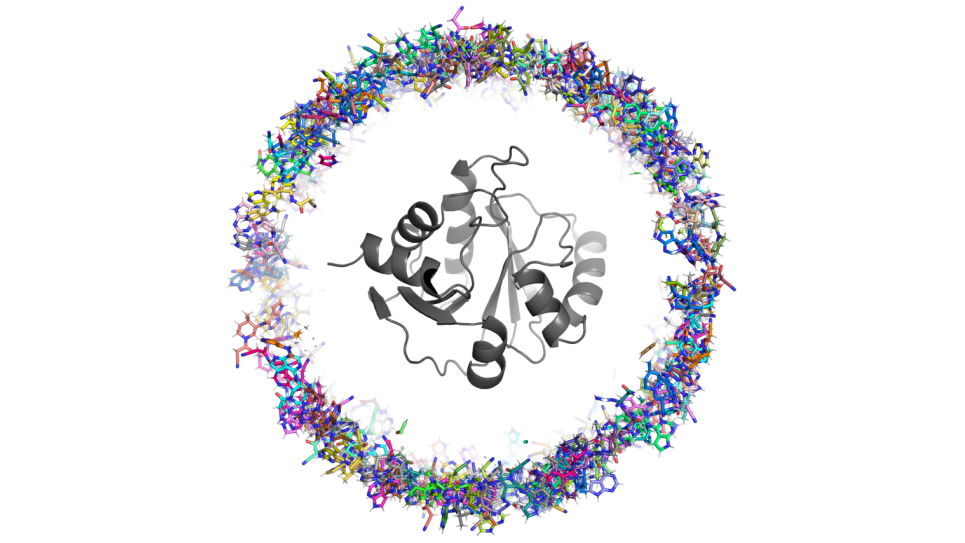
“Ensemble of uniformally distributed initial positions of a candidate repurposed drug prior to its interaction with the COVID-19 protein target ADRP, performed over many long-time molecular dynamics simulations run on Summit at OLCF.”
Credit: Art Hoti, Aghastya Bhati, and Peter Coveney, University College London