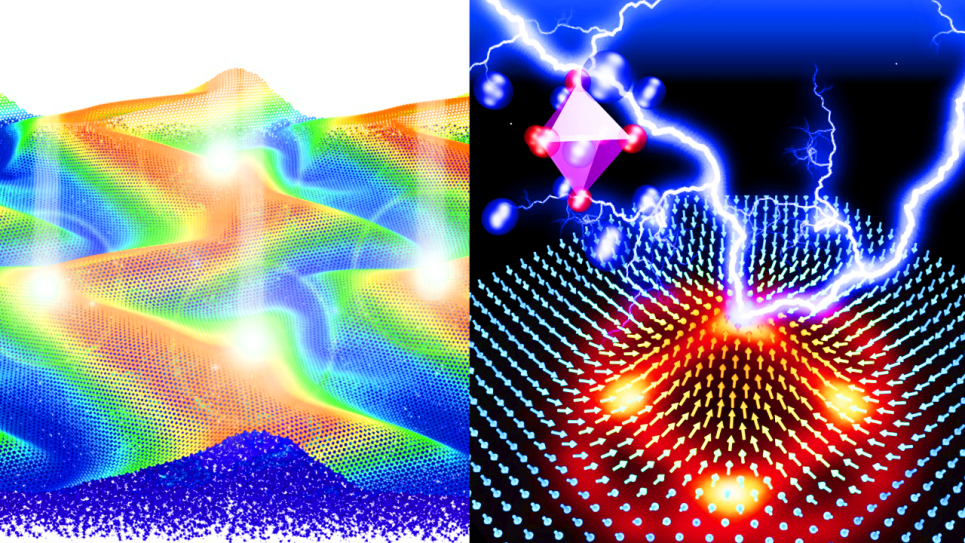
Metascalable Layered Materials Genome
This project will advance layered materials genome (LMG). Functional layered materials (LM) will dominate materials science in this century. The attractiveness of LMs lies not only in their outstanding electronic, optical, magnetic and chemical properties, but also in the possibility of tuning these properties in desired ways by building van der Waal (vdW) heterostructures composed of unlimited combinations of atomically thin layers. The researchers will perform 10^5-atom nonadiabatic quantum molecular dynamics (NAQMD) and 10^10-atom reactive molecular dynamics (RMD) simulations on Aurora for computational synthesis and characterization of LMs. The unprecedented simulations will (1) guide the synthesis of stacked LMs by chemical vapor deposition (CVD), exfoliation and intercalation, and (2) discover function-property-structure relationships in LMs with a special focus on far-from-equilibrium electronic processes.
This project will advance layered materials genome (LMG). Functional layered materials (LM) will dominate materials science in this century. The attractiveness of LMs lies not only in their outstanding electronic, optical, magnetic and chemical properties, but also in the possibility of tuning these properties in desired ways by building van der Waal (vdW) heterostructures composed of unlimited combinations of atomically thin layers. The researchers will perform 10^5-atom nonadiabatic quantum molecular dynamics (NAQMD) and 10^10-atom reactive molecular dynamics (RMD) simulations on Aurora for computational synthesis and characterization of LMs. The unprecedented simulations will (1) guide the synthesis of stacked LMs by chemical vapor deposition (CVD), exfoliation and intercalation, and (2) discover function-property-structure relationships in LMs with a special focus on far-from-equilibrium electronic processes.
The metascalable (or “design once, scale on new architectures”) simulation approach used here achieves portable performance on current and future computing platforms based on a novel divide-conquer-“recombine” (DCR) algorithmic framework for (1) lean divide-and-conquer density functional theory (LDC-DFT) for NAQMD simulations with minimal O (N) prefactor, and (2) extended Lagrangian RMD (XRMD) to eliminate the speed-limiting charge iterations in RMD simulations . Key to metascalability is global-local separation achieved by our globally scalable /reproducible and locally fast (GSLF) solvers based on (1) a new scalable and reproducible global summation method, and (2) fast shift-collapse (SC) computation of local n-tuples.