Scaling Genomic Variant Callers to Leadership-Class Systems: A Collaboration Between VA-MVP and DOE
This project builds upon the strategic partnership between the US Department of Veterans Affairs (VA) and DOE in leveraging DOE computing capabilities to enhance the health outcomes of veterans through the MVP-CHAMPION (Million Veteran Program Computational Health Analytics for Medical Precision to Improve Outcomes Now) project.
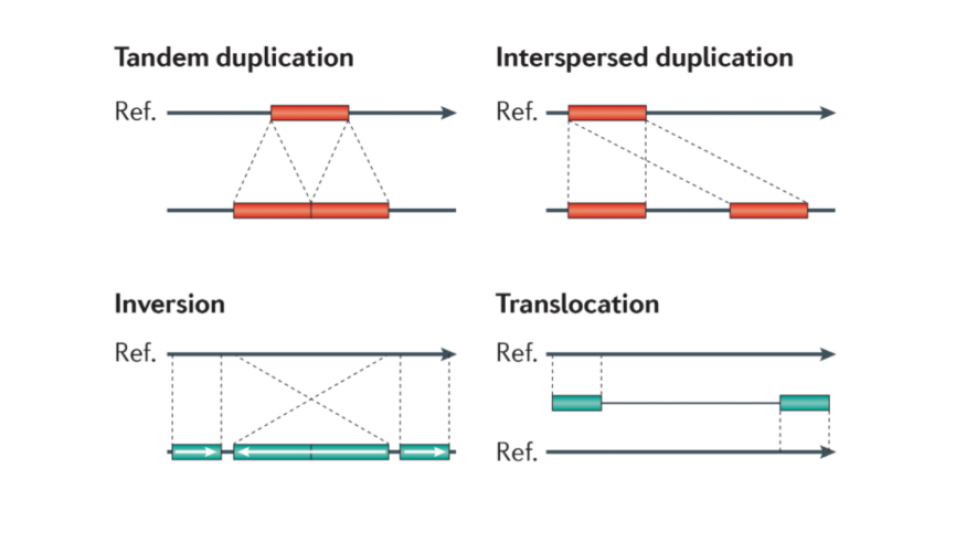
The human genome, which consists of 23 pairs of chromosomes containing over 3 billion nucleotide base pairs (A-T and C-G pairs) encodes the information for the formation of proteins that drive all life processes. Conditions like autism, hemophilia, schizophrenia, cardiovascular diseases, Huntington’s disease, cancer and Alzheimer’s, among many others, can be caused by malfunctioning proteins that are in turn related to the large-scale changes in the human genome referred to as “structural variations'' (SVs). SVs are difficult to identify due to their complex nature as well as some inherent limitations of genome sequencing and mapping techniques. This study will identify and quantify SVs from the 100,000 whole human genomes sequenced using Next Generation Sequencing (NGS) technique as part of the Million Veteran Program (MVP). NGS platforms create multiple copies of the 3 billion base pair long genome and split them into tens of millions of short pieces (referred to as “reads”) for the ease of accurate identification of the bases. For this work, these randomly located reads will be rearranged for each of the genomes by “mapping” them to the human reference genome, and then identification and characterization of SVs will be done using AI-assisted and traditional methods. The Aurora supercomputer will be used to carry out these computationally intensive tasks as well as to generate synthetic genomes and train the AI-assisted models for improved accuracy.
The goals of this project align with the US Department of Energy's (DOE) mission to promote transformative growth in scientific research using supercomputers and AI. It builds upon the strategic partnership between the US Department of Veterans Affairs (VA) and DOE in leveraging DOE computing capabilities to enhance the health outcomes of veterans through the MVP-CHAMPION (Million Veteran Program Computational Health Analytics for Medical Precision to Improve Outcomes Now) project. By cataloging SVs from over hundreds of thousands of genomes available from various sequencing projects, this project will create one of the largest databases of SVs in the world. The inclusion of participants from diverse racial backgrounds further enhances the quality and representativeness of the data. Curated SVs from such a large and diverse population will enhance the statistical power of genome-wide association studies (GWAS), allowing for the identification of beter correlation between large genomic variations and phenotypes that will greatly facilitate the early detection of variants responsible for various conditions, and allow quicker identification of therapeutic targets for precision medicine.