Inside the Skull
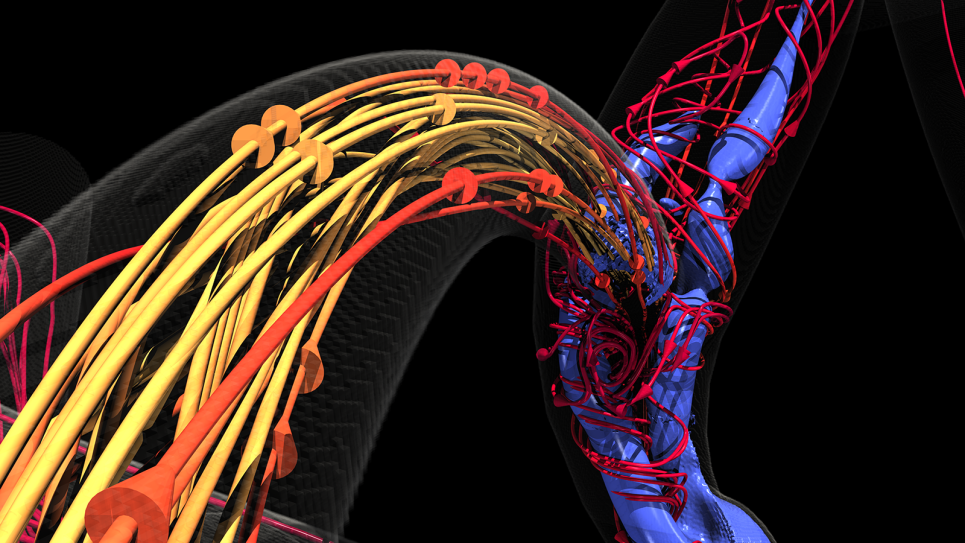
Blood supply to the brain relies on a highly complex system where, at any point, an aneurysm may occur. Angiograms can show the presence of aneurysms and clots but don’t necessarily reveal their cause – the interactions among and between platelets and red and white blood cell and the endothelial cells that line blood vessels. When injured, endothelial cells trigger platelet aggregation, leading to a clot.
Conventional imaging also doesn’t show the big picture of blood flow throughout the brain – where blood is coming from and where it’s going. Precisely understanding these elements at the level of both gross blood flow and its microscopic properties would greatly improve neurosurgeons’ ability to predict when and where an aneurysm might rupture and when to operate.
High-performance computing (HPC) shows promise for simulating and visualizing brain blood flow at multiple scales. Paving the way is a team led by Leopold Grinberg, senior research associate in the Division of Applied Mathematics at Brown University. Other researchers include Brown’s George Em Karniadakis, Argonne National Laboratory’s (ANL) Joseph A. Insley, Vitali Morozov, Michael E. Papka and Kalyan Kumaran, and Dmitry Fedosov of the Institute of Complex Systems (ICS) in Jülich, Germany.
In 2006, Grinberg began developing arithmetical and software methods capable of simulating 3-D blood flow in complex arterial networks. “The methodology I started to build was based on functional decomposition, with many tasks assigned to different groups of processors, plus multilevel communicating interfaces capable of connecting the data computed by different tasks and synchronizing the processors assigned to different tasks,” Grinberg says.
Working on three fronts – parallel computing, mathematical algorithms and visualization tools – the team from Brown and ANL received 50 million processor hours on Intrepid, Argonne’s IBM Blue Gene/P, HPC, and 23 million hours on Jaguar, the Cray XT system at Oak Ridge National Laboratory (ORNL). The allocations were granted through the Department of Energy’s Innovative and Novel Computational Impact on Theory and Experiment (INCITE) program. The researchers also had access to Jugene, ICS’s Blue Gene/P, which runs at a peak speed of 1 petaflops – a quadrillion calculations per second – and is almost twice the size of Intrepid.
As they reported in November at the SC11 high-performance computing conference in Seattle, Grinberg and colleagues have created what they think is the first truly multiscale simulation and visualization of an actual biological system. The team’s ambitious target was brain blood flow, the most complex arteriovenous system in the human body. A paper describing the research was one of five finalists for the prestigious 2011 Gordon Bell Prize, which recognizes outstanding results in the application of parallel computing to practical scientific problems.
The geometry and some blood flow conditions were derived from a real patient. Blood properties (viscosity, for example) were based on data for an average person. With this information, the researchers used ANL and ORNL computers to refine the code to run successfully on HPCs. “A simulation run on an HPC such as Blue Gene/P can involve a very large number of cores – more than 131,000 – and take almost 25 hours nonstop to complete for just one heart beat,” Grinberg says. “These simulations take a long time to run even on an HPC, and you don’t want the code to fail after only a minute or one hour.”
These are monumental computations. Modeling just 1 cubic millimeter of human blood involves 1 billion to 2 billion dissipative particle dynamics, or DPDs. “DPDs involve a technique to simulate the dynamics of clusters of particles, instead of each particle individually,” Morozov says. “In other words, each DPD particle represents a set of many particles and is treated as if it is a big individual particle.”
From centimeters to microns
“We have been able to resolve simulations at multiple scales, not just one as others have done, and in parallel fashion,” Grinberg says. In other words, the team’s methods make it possible for the first time to simultaneously see a snapshot of blood flow throughout the entire brain on a scale that ranges from as large as 4 to 5 centimeters down to less than a micron.
“This is a truly unique advance in computational science,” Morozov says. “Many different approaches have been suggested in the past, but none of them really succeeded in getting practical results.”
Grinberg expects that the methods the group developed also will be useful for simulating other subjects, such as crack propagation in aircraft surfaces at the molecular level. “This type of research overlaps very well with what we are trying to do, but we are working on a different problem: We model blood cells as they aggregate and interact on the larger scales.”
So what’s the outlook for multiscale simulations in medicine? Grinberg says researchers must increase model complexity and devise codes that are fast enough to perform longer multiscale simulations than now possible. Hospitals and other medical institutions are turning more and more to computing to model different problems and to predict the consequences of surgery.
But until exaflop computers – roughly a thousand times more powerful than today’s best machines – arrive in a decade or more, it’s unlikely front-line doctors can use real-time simulations and visualizations like the relatively simple, early ones Grinberg and others are developing. Nonetheless, Grinberg predicts, future doctors will receive training that incorporates HPCs in their practices, and hospitals will staff specialists who can tap into computing resources at national centers.
Says Ginberg, “The day will come when we’ll be able to develop software that can enable very sophisticated models and simulations – how many more types of cells interact, for example – and physicians will have much better understanding of how to treat their patients.”